The Bfiat package: modelling trawling effects in the Dutch part of the Northsea
Karline Soetaert and Olivier Beauchard
Source:vignettes/MWTL.Rmd
MWTL.RmdAbstract
The impact of bottom trawling on benthic life in the Dutch part of the Northsea is estimated. We use monitoring data from 103 stations in the area (MWTL data set), and impose fishing intensities estimated for each of these stations.Aim.
We use functions from the Btrait and Bfiat R-packages to estimate the impact of bottom fisheries on sediment ecosystem functioning in the Dutch part of the Northsea.
We use three sets of data:
- in situ density data from a particular site.
- species trait information, including:
- the age at maturity of the species, and
- the depth of occurrence in the sediment.
- fishing intensity and gear penetration depths in the sites
We use the MWTL macrofauna density data that have been compiled in the frame of the EMODnet biology project, (https://www.emodnet-biology.eu). These data are available from the R-package Btrait (Soetaert and Beauchard, 2024a).
The NIOZ trait database is used for the species traits (Beauchard et al., 2021, 2023). The trait dataset is also in the R-package Btrait.
Software.
The fishing impact models run in the open source framework R (R core team, 2025) and have been implemented in the Bfiat R-package (Soetaert and Beauchard, 2024b).
The R-package Btrait (Soetaert and Beauchard, 2022) contains functions to work on density and trait datasets.
The packages Btrait and Bfiat are available on github (https://github.com/EMODnet/Btrait and https://github.com/EMODnet/Bfiat).
The fisheries impact model
The fisheries impact model is based on the logistic growth equation (Verhulst 1845) that describes how species density or biomass evolves over time. The effect of fishing on the population is included as a series of events, that instantaneously decimate population sizes, while in between these events the populations recover.
The model thus consists of two parts: (continuous) logistic growth in between fishing events and an instantaneous depletion during fishing.
Logistic growth
The differential equation is, for t in between and : where is time, is the density of species at a particular time, (units of density) is the carrying capacity of species , is the logistic growth parameter (units [/time]).
Instantaneous trawling
During trawling at times , density changes instantaneously:
where and are the density immediately after and before trawling occurs at time respectively, and is the species-specific and trawl-specific depletion fraction.
Trawling intensity
The time in between trawling can be inferred from the fishing intensity, expressed as the annual swept area ratio (SAR).
SAR is the cumulative area contacted by a fishing gear over one year and per surface area. It has units of , or .
From the swept area ratio (hereafter denoted as ), we calculate the average time in between fishing events () as:
Data requirements
Central in the impact analysis is the use of a combination of data sets:
Community composition
Benthic biological data to be used for fishing assessment should comprise at least density data, and preferentially also biomass of individual species.
The densities or biomasses are used to estimate the “carrying capacity” of the species at a particular site; this is the abundance the species attains at steady-state, when undisturbed (i.e. in the absence of fishing).
Taxonomy
To assign biological traits to the taxa in the community, it is convenient to know their taxonomic position. Thus it will be possible to derive trait characteristics for taxa that are not documented in the trait database, based on their taxonomic relationships.
Trait characteristics
The following species traits are used to estimate the fishing impact on species density:
- The age at maturity of species is used to estimate their “intrinsic rate of natural increase” ().
- The depth at which species live determines their vulnerability to bottom trawling: it is used to derive the model’s “depletion parameter” ().
Fishing intensity
The model also needs the fishing intensity for a certain area, for instance expressed as the annual swept area ratio (SAR); this is the cumulative area contacted by a fishing gear over one year and per surface area.
From the swept area ratio (hereafter denoted as , units ), we calculate the time in between fishing events () as .
Data availability
Community composition
For the subsequent analyses, we use macrofauna density data that have been compiled in the frame of the EMODnet biology project, (https://www.emodnet-biology.eu).
The monitoring data from the Dutch part of the North sea (called the MWTL monitoring data) comprise both species densities and biomass for 103 stations, that were sampled during 19 years, extending from 1995 till 2018.
Sampling was performed yearly at first, then less frequently. Calculations in this document are based on yearly averages.
The MWTL data are available from the R-package Btrait, and are in a list called MWTL (within R, type: ?MWTL to open the help file for these data).
The composition of the MWTL data is in Appendix 1.
| title | originator | implemented |
|---|---|---|
| The MWTL Northsea macrobenthos data (1995 - 2018) | Ministerie van Infrastructuur en Milieu, Rijkswaterstaat Centrale Informatievoorziening (RWS, CIV) | Karline Soetaert, in the frame of the EMODnet biology project |
| station | date | year | taxon | density | biomass |
|---|---|---|---|---|---|
| BREEVTN02 | 27-06-1995 | 1995 | Bathyporeia guilliamsoniana | 14.6 | 0.0044 |
| BREEVTN02 | 27-06-1995 | 1995 | Callianassa | 58.5 | 1.3986 |
| BREEVTN02 | 27-06-1995 | 1995 | Chaetozone setosa | 14.6 | 0.0073 |
Taxonomic data
There are 400 different taxa in the MWTL data.
The taxonomic information for all taxa, has been derived from the WoRMS database and is available from package Btrait, in dataset Taxonomy:
| taxon | genus | family | order | class | phylum | AphiaID |
|---|---|---|---|---|---|---|
| Abietinaria | Abietinaria | Sertulariidae | Leptothecata | Hydrozoa | Cnidaria | 117225 |
| Abludomelita | Abludomelita | Melitidae | Amphipoda | Malacostraca | Arthropoda | 101665 |
| Abludomelita obtusata | Abludomelita | Melitidae | Amphipoda | Malacostraca | Arthropoda | 102788 |
Trait data
We use the traits compiled by NIOZ (called Traits_nioz) for age at maturity, and living depth. These trait data (mainly on species level) were compiled by Beauchard et al. (2021) and Beauchard et al. (2023). This database records 32 different traits from 281 taxa.
All traits are fuzzy-coded. The data structure is in appendix 2.
| taxon | ET1.M1 | ET1.M2 | ET1.M3 | ET1.M4 |
|---|---|---|---|---|
| Abludomelita | 0.5 | 0.5 | 0.0 | 0 |
| Abludomelita obtusata | 0.5 | 0.5 | 0.0 | 0 |
| Abra alba | 0.0 | 0.5 | 0.5 | 0 |
The figure below shows the position and area of the MWTL stations, and the swept area ratios (sar) that were derived from OSPAR (ICES 2018).
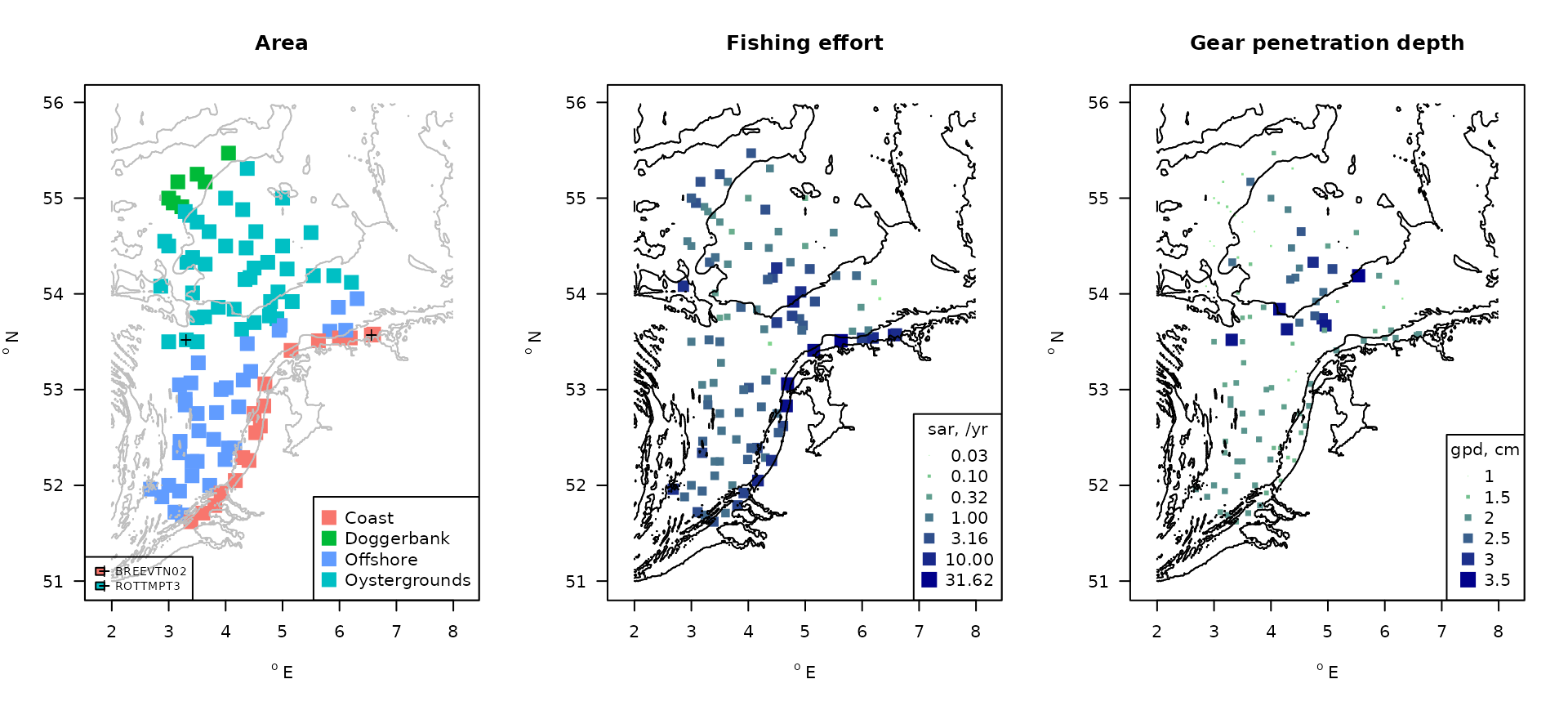
FIG: MWTL sampling stations with indication of the main areas, fishing intensity and gear penetration depth. ‘+’ denote the two stations that will be showcased.
Model steps
The model is applied in several steps:
- the taxon densities per station are prepared, and a list of taxa in the data is generated,
- the traits recording age at maturity and the depth distribution for each taxon are extracted,
- the fishing parameters for each station are extracted
- the parameter values are estimated based on the traits, fishing parameters, and taxon density
- the model is run
The MWTL density
The MWTL data are gathered over several years, and are in data.frame MWTL$density. We calculate the density for all stations (descriptor) and taxa, averaged over all years, using Btrait function get_density.
MeanMWTL <-
get_density(data = MWTL$density,
descriptor = station,
taxon = taxon,
value = density,
averageOver = year)The taxa whose traits need to be found are extracted:
Extracting taxon traits
Taxon traits that we need are in the nioz trait database (Traits_nioz), from R-package Btrait. From this database, we use:
- the depth distribution (fractional occurrence in each depth layer),
- the age at maturity of a species
Substrate depth distribution
Depth distribution is a fuzzy-coded trait in the trait database. For use in the model we need to keep it fuzzy coded.
The meaning of the nioz traits is stored in its metadata. We look for the trait names that relate to depth distribution:
meta <- metadata(Traits_nioz) # description of trait modalities
meta.D <- subset(meta, # names of trait modalities for substratum depth
subset = (trait == "Substratum depth distribution"))| colname | trait | modality | indic | value | score | units |
|---|---|---|---|---|---|---|
| ET1.M1 | Substratum depth distribution | 0 | 1 | 0.0 | 1.00 | cm |
| ET1.M2 | Substratum depth distribution | 0-5 | 1 | 2.5 | 0.75 | cm |
| ET1.M3 | Substratum depth distribution | 5-15 | 1 | 10.0 | 0.50 | cm |
| ET1.M4 | Substratum depth distribution | 15-30 | 1 | 22.5 | 0.25 | cm |
| ET1.M5 | Substratum depth distribution | >30 | 1 | 30.0 | 0.00 | cm |
We then extract the depth traits for each taxon in the MWTL density data set. As not all species in the MWTL dataset have traits assigned, we use the taxonomic information (in dataset Taxonomy) to estimate the traits also for unrecorded species.
traits.D <- get_trait(
taxon = MWTLtaxa,
trait = Traits_nioz[, c("taxon", meta.D$colname)],
taxonomy = Taxonomy)
colnames(traits.D)[-1] <- meta.D$modality # suitable column names| taxon | 0 | 0-5 | 5-15 | 15-30 | >30 |
|---|---|---|---|---|---|
| Abludomelita obtusata | 0.5 | 0.5 | 0.0 | 0 | 0 |
| Abra alba | 0.0 | 0.5 | 0.5 | 0 | 0 |
| Abra nitida | 0.0 | 1.0 | 0.0 | 0 | 0 |
Of the 400 taxa in the MWTL dataset, only 209 taxa are documented in the trait database. The traits for the remaining taxa were estimated based on taxonomic closedness. Of these taxa, only 8 could not be assigned a trait. These were: Ascidiacea, Chaetognatha, Enteropneusta, Entoprocta, Nematoda, Nemertea, Platyhelminthes, Priapulida. As these taxa are not true macrofauna, they can be removed from the dataset.
Notrait <- attributes(traits.D)$notrait
MWTLtaxa <- MWTLtaxa [! MWTLtaxa %in% Notrait]
MeanMWTL <- MeanMWTL [! MeanMWTL$taxon %in% Notrait, ]Age at maturity
We perform a similar procedure to extract the age at maturity for each taxon in the density data. The age at maturity is estimated as the average of the fuzzy values.
We first find the relevant trait names in the trait database:
meta.A <- subset(meta,
subset = (trait == "Age at maturity"))| colname | trait | modality | indic | value | score | units |
|---|---|---|---|---|---|---|
| RT8.M1 | Age at maturity | <1 | 8 | 0.5 | 0.0 | year |
| RT8.M2 | Age at maturity | 1-3 | 8 | 2.0 | 0.5 | year |
| RT8.M3 | Age at maturity | 3-5 | 8 | 4.0 | 1.0 | year |
We then calculate the average age at maturity. To calculate the average, we pass the trait_class and trait_score while extracting the traits.
traits.A <- get_trait(
taxon = MWTLtaxa,
trait = Traits_nioz[, c("taxon", meta.A$colname)],
trait_class = meta.A$trait,
trait_score = meta.A$value,
taxonomy = Taxonomy)| taxon | Age.at.maturity |
|---|---|
| Abludomelita obtusata | 0.5 |
| Abra alba | 0.5 |
| Abra nitida | 0.5 |
Merged trait-density data.frame
We merge these two trait datasets in one data.frame, merging by the taxon.
TraitsAll <- merge(x = traits.A,
y = traits.D,
by = "taxon")and then combine it with the species density data
MWTLtrait <- merge(x = MeanMWTL,
y = TraitsAll,
by = "taxon")Fishing parameters
Each station has a specific fishing intensity (swept area ratio, sar), and mean gear penetration depth (gpd). This information is kept in data.frame MWTL$abiotics (from R-package Btrait).
The MWTL data.frame are merged with the swept area ratio, and with the gear penetration depth, merging on column station.
Parameter values
Intrinsic rate of natural increase
The intrinsic rate of natural increase is specific for a species, and can be estimated using function par_r. This parameter is added to the MWTL data.frame.
MWTLtrait$r <- par_r(age_at_maturity = MWTLtrait$Age.at.maturity)Depletion fraction d
The depletion rate depends on the depth occurrence of the species (fDepth), and on the gear penetration depth (gpd).
The latter differs among stations, so the species depletion fraction will vary with the station.
We also need to input the upper position of each depth layer (uDepth).
Carrying capacity K
Te carrying capacity is estimated from the model itself. We pass the species density, the fishing intensity (sar), the rate of increase (r), and the depletion fraction (d):
MWTLtrait$K <- par_K(
density = MWTLtrait$density,
sar = MWTLtrait$sar,
r = MWTLtrait$r,
d = MWTLtrait$d
)The data that we will use for fishing impact assessment is:
| station | taxon | r | d | K | sar | gpd |
|---|---|---|---|---|---|---|
| BREEVTN02 | Ophiura | 0.96 | 0.16 | 2.87 | 1.42 | 2.95 |
| BREEVTN02 | Sthenelais limicola | 0.73 | 0.14 | 5.64 | 1.42 | 2.95 |
| BREEVTN02 | Diplocirrus glaucus | 1.89 | 0.11 | 6.23 | 1.42 | 2.95 |
Species for which the model does not find a solution
These are species where the depletion is too high and/or the intrinsic rate of increase is too low, so that it cannot persist in the environment under the current fishing pressure. This causes an anomalously high carrying capacity compared to observed density. Such occurrences are removed from the data set.
MWTLtrait$DK <- MWTLtrait$density/MWTLtrait$K
ii <- which(is.na(MWTLtrait$DK))
NAS <- subset(MWTLtrait, subset = is.na(DK))
MWTLtrait <- MWTLtrait[-ii,]There are 16 such species, the highest observed density being17.4415692. These taxa are removed from the analysis.
TT <- data.frame(table(NAS$taxon))
TM <- aggregate(NAS$density, list(NAS$taxon), FUN=max)
TT <- merge(TT, TM, by = 1)
names(TT) <- c("taxon", "occurrences", "max density")
knitr::kable(TT, caption = "Taxa for which the model could not be solved")| taxon | occurrences | max density |
|---|---|---|
| Amphiura filiformis | 1 | 0.6747638 |
| Astropecten | 2 | 11.5504579 |
| Crangon crangon | 1 | 0.6747638 |
| Cucumaria | 1 | 0.6842105 |
| Ensis ensis | 1 | 2.4416667 |
| Eteone longa | 1 | 2.9852901 |
| Eudorella truncatula | 1 | 3.4690862 |
| Kurtiella bidentata | 1 | 7.0395316 |
| Ophiura | 2 | 4.0242915 |
| Paraleptopentacta elongata | 1 | 1.4431849 |
| Pholoe baltica | 1 | 5.4170040 |
| Pholoe minuta | 2 | 13.2036283 |
| Spisula subtruncata | 1 | 17.4415692 |
Running the fishing model
We run the model for 8255 species-station occurrences. The model is run for 30 years, of which 20 years of fishing (and thus 10 years where fishing stopped).
We provide taxon names to label the output columns
times <- seq(from = 0, to = 30, length.out = 3000)
system.time(
dens <- density_perturb(parms = MWTLtrait,
times = times,
taxon_names = paste(MWTLtrait$station,
MWTLtrait$taxon, sep = "_"),
tend_perturb = 20)
)## user system elapsed
## 0.552 0.067 0.619The result is a matrix with 3000 rows and 8256 columns, the first of which contains the time, followed by the density trajectories of all species, and for all stations.
| times | BREEVTN02_Ophiura | BREEVTN02_Sthenelais limicola | BREEVTN02_Diplocirrus glaucus |
|---|---|---|---|
| 0.00 | 2.87 | 5.64 | 6.23 |
| 0.01 | 2.87 | 5.64 | 6.23 |
| 0.02 | 2.87 | 5.64 | 6.23 |
Summaries
The total density per station is calculated as rowSums per station.
Dens <- data.frame(dens[, -1]) # remove first column
Total <- sapply(split.default(Dens, MWTLtrait$station), # split in stations
FUN = rowSums, na.rm = TRUE) # take row sumsThen relative values are calculated by dividing all columns (MARGIN = 2) by their maximum (STATS).
Some results
We show the results for the 20 most abundant species in two stations.
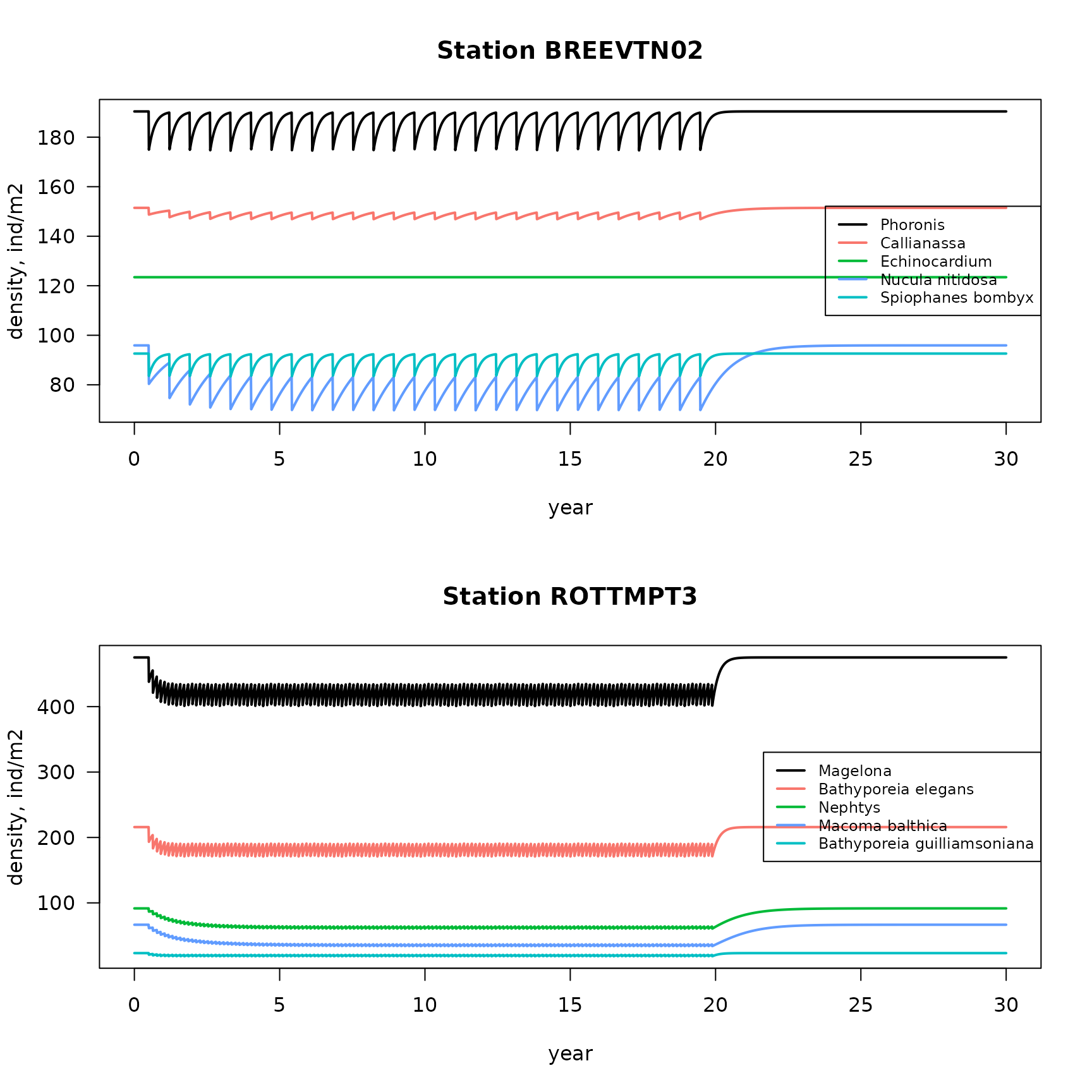
FIG: Density trajectories of 5 most abundant species from two stations
The average density reduction due to fishing in each of the stations is estimated:
# fishing reduction for all species
DDrel <- apply(X = dens[, -1],
MARGIN = 2,
FUN = function(x) 1-min(x)/max(x))
# averaged per station
DDstat <- aggregate(x = list(p = DDrel),
by = list(station = MWTLtrait$station),
FUN = mean)
MWTLstat <- merge(MWTL$stations, DDstat, # add station coordinates
by = "station")
with(MWTLstat,
map_MWTL(x = x, y = y, colvar = p,
main = "Fractional reduction ", clab = "-",
col = Colramp, cex = 2, pch = 15, log = "",
type = "legend")
)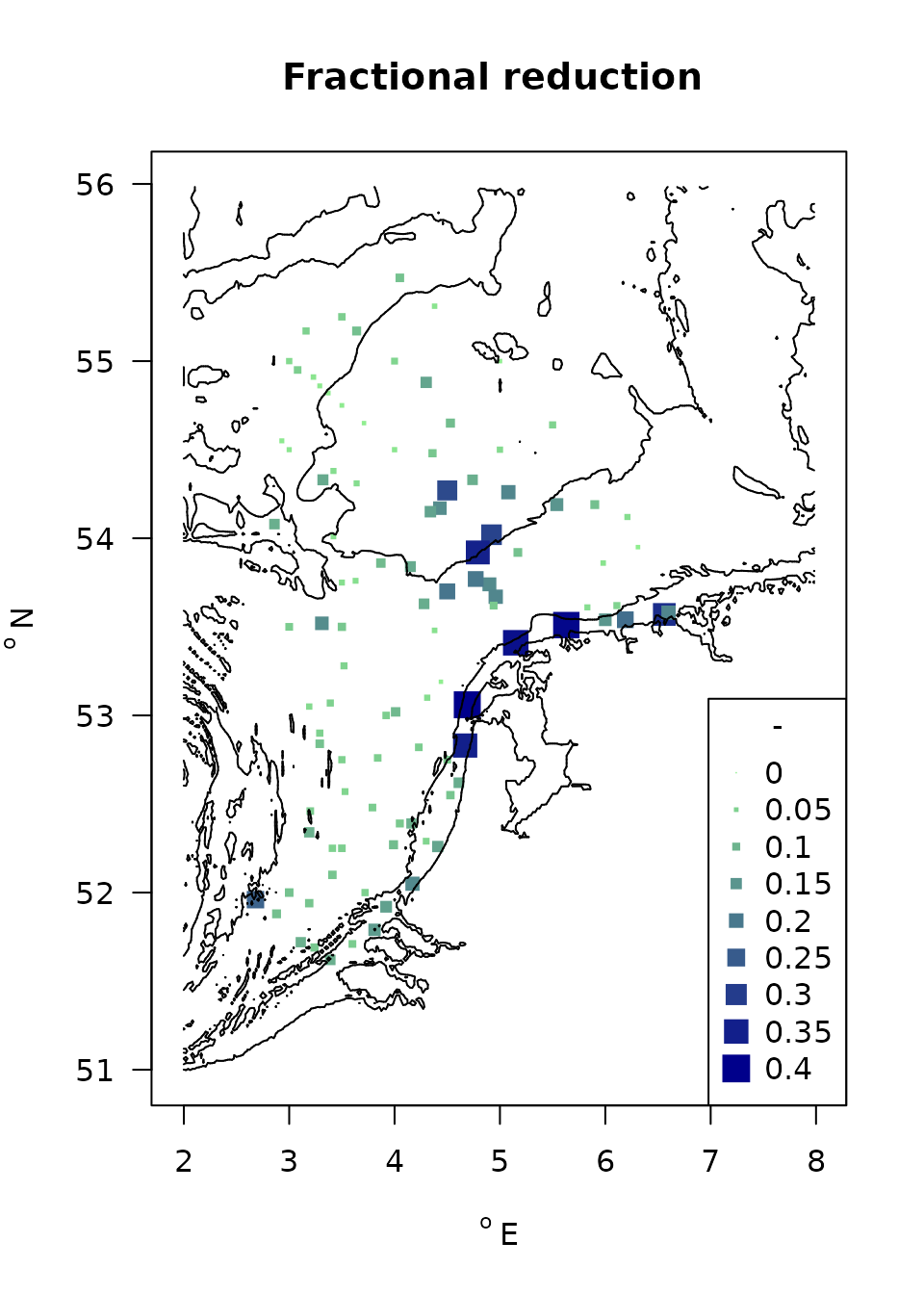
FIG: Average fishing impacts for the MWTL stations
Appendix
Structure of the MWTL data
The MWTL list contains several data.frames. We will use mainly the data.frames called density, stations, and abiotics (for fishing intensities). The structure of these data is:
| name | description | format |
|---|---|---|
| x | degrees longitude | WGS84 |
| y | degrees latitude | WGS84 |
| name | description | units |
|---|---|---|
| station | station name | |
| date | sampling date, a string | |
| taxon | taxon name, checked by worms, and adjusted | |
| density | species total density | individuals/m2 |
| biomass | species total ash-free dry weight | gAFDW/m2 |
| taxon.original | original taxon name |
| name | description | units |
|---|---|---|
| DRB | swept area ratio for dredge | m2/m2/year |
| OT | swept area ratio for otter trawl | m2/m2/year |
| SN | swept area ratio for seines | m2/m2/year |
| TBB | swept area ratio for beam trawl | m2/m2/year |
| sar | swept area ratio (fisheries), DRB +OT +SN +TBB | m2/m2/year |
| subsar | swept area ratio (fisheries) > 2cm | m2/m2/year |
| gpd | gear penetration depth | cm |
Structure of the Trait data
| colname | trait | modality | indic | value | score | units |
|---|---|---|---|---|---|---|
| ET1.M1 | Substratum depth distribution | 0 | 1 | 0.0 | 1.00 | cm |
| ET1.M2 | Substratum depth distribution | 0-5 | 1 | 2.5 | 0.75 | cm |
| ET1.M3 | Substratum depth distribution | 5-15 | 1 | 10.0 | 0.50 | cm |
| ET1.M4 | Substratum depth distribution | 15-30 | 1 | 22.5 | 0.25 | cm |
| ET1.M5 | Substratum depth distribution | >30 | 1 | 30.0 | 0.00 | cm |
| ET2.M1 | Biodiffusion | Null | 2 | 0.0 | 0.00 | - |
References
Beauchard O, Brind’Amour A, Schratzberger M, Laffargue P, Hintzen NT, Somerfield PJ, Piet G (2021) A generic approach to develop a trait-based indicator of trawling-induced disturbance. Mar Ecol Prog Ser 675:35-52. https://doi.org/10.3354/meps13840
Beauchard, O., Murray S.A. Thompson, Kari Elsa Ellingsen, Gerjan Piet, Pascal Laffargue, Karline Soetaert, 2023. Assessing sea floor functional diversity and vulnerability. Marine Ecology Progress Series v708, p21-43, https://www.int-res.com/abstracts/meps/v708/p21-43
ICES Technical Service, Greater North Sea and Celtic Seas Ecoregions, 29 August 2018 sr.2018.14 Version 2: 22 January 2019 https://doi.org/10.17895/ices.pub.4508 OSPAR request on the production of spatial data layers of fishing intensity/pressure.
R Core Team (2024). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Soetaert Karline, Olivier Beauchard (2024). Btrait: Working with Biological density, taxonomy, and trait composition data. R package version 1.0.1.
Soetaert Karline, Olivier Beauchard (2024). Bfiat: Bottom Fishing Impact Assessment Tool. R package version 1.0.1.
Verhulst, Pierre-François (1845). “Recherches mathématiques sur la loi d’accroissement de la population” [Mathematical Researches into the Law of Population Growth Increase]. Nouveaux Mémoires de l’Académie Royale des Sciences et Belles-Lettres de Bruxelles. 18: 8.